The Bagheri Lab integrates experimental data with computational strategies to elucidate fundamental properties governing intracellular dynamics and intercellular regulation. Our group is highly collaborative and integrates a diverse array of research interests. We take on grand challenges spanning complex dynamics of cell populations, to experimental design and tool development. A common thread that persists among our projects involves elucidating, predicting, and ultimately controlling biological response, particularly in context of disease. When the regulation, or control, of biological function fails, people can manifest a variety of illnesses including cancer and autoimmune disease.
Ongoing collaborative projects span three major thematic areas: emergent dynamics, machine learning, and network theory.
Emulation of computationally expensive models towards statistically rigorous analyses
Jason Y. Cain | Jacob I. Evarts
machine learningemergent dynamics
Computational models aim to include enough biological detail to highlight important phenomena, but introducing too much detail can incur prohibitive computational costs. Running exhaustive simulations required for statistically rigorous analyses quickly becomes difficult or intractable. One possible alternative is to run exhaustive simulations on a separate model, trained to emulate the behavior of the original simulation. We identify the appropriate learning goals to replicate the emergent function of interest as a key step to stengthening insights we can extract from agent-based models.
Hypoxia-angiogenesis feedback loop and tumor development
emergent dynamicsnetwork theory
Hypoxia, a hallmark of most solid tumors, is highly associated with poor patient outcomes. Hypoxic cells secrete angiogesis factors, providing cytokines to support blood vessel development and nutrient distribution. This relationship results in hypoxia being both a driver and a consequence of cancer progression. An accurate, robust model of the multiscale relationship between hypoxia and angiogenesis is critical to guiding interventions. Agent-based models are an intuitive solution to capture the emergent dynamics of this niche as well as to model interventions that target hypoxia or angiogenesis.
Lateral root development
emergent dynamics
Root system architecture is a major determinant of plant fitness. The ability to precisely control root system architecture would enable farmers to optimize plant fitness for field conditions, maximizing production efficiency. We aim to interrogate mechanisms underlying cellular decision making during lateral root development in dicot plants using an agent-based model. We will use this model to identify control inputs and perturbations that allow for the precise control of lateral root formation.
Decomposing information sources in simulations
emergent dynamics
Agent-based models are able to capture spatial and temporal changes in an environment. Complex behaviors can often emerge out of these systems despite simple rulesets and actions. We aim to decompose the information sources across time to better understand these unexpected phenomena. We will use this to make better predictions about a system's behavior.
Multiscale, multiclass models of cell populations
emergent dynamics
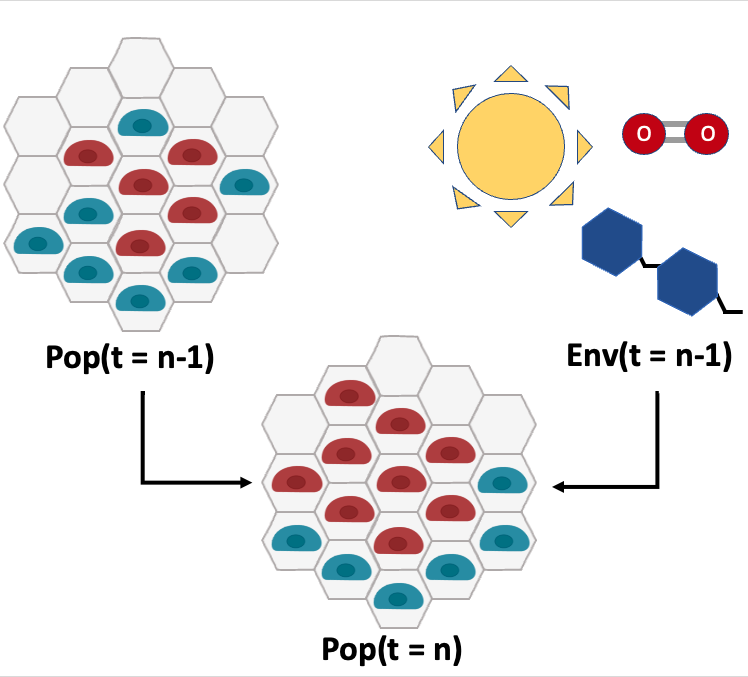
To effectively understand and predict cell population responses to intrinsic perturbations and extrinsic intervention, the integration of intracellular signaling with intercellular dynamics is necessary. Through agent-based models, we simulate and predict the behavior of heterogeneous cell populations. Our in silico framework (developed by Jessica S. Yu) effectively elucidates how changes at the subcellular level emerge into varied population level responses.
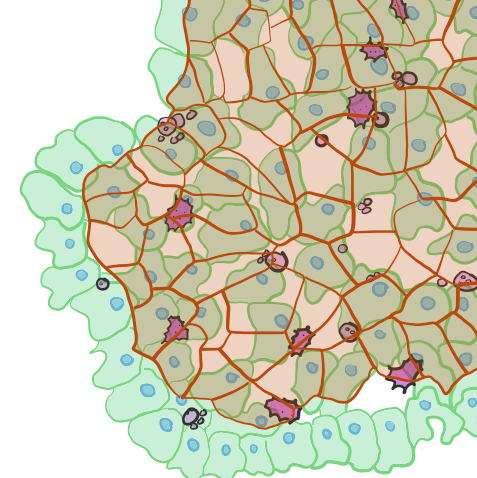
Wound healing in complex epithelia
emergent dynamicsnetwork theory
Cells in a tissue respond autonomously to their environment and to perturbations without a central regulator. Their collective, decentralized behavior gives rise to emergent cell population/tissue dynamics. The underlying cell level rules that govern individual cell behavior and subsequent emergent dynamics of a tissue during wound healing are largely unknown in complex epithelia. We are integrating ex vivo zebrafish skin tissue microscopy with high spatio-temporal resolution along with graph theory and agent-based modeling to uncover rules that drive wound healing.